Molecular diagnostic issues
- To submit samples for one or more of the below analyses, please use the CHUV-IMU order form No 151 (provided upon request to 021-3144107).
- For the diagnosis of HBV resistance and/or genotyping, please make sure that the HBV DNA is positive, and complete the clinical details field of the CHUV-IMU order form No 151.
- Adequate blood samples are either EDTA-K blood or plasma (5-10 ml, priority mail, ambient temperature). Serum has also been used succesfully for HDV and HEV RNA detection.
- CSF can be evaluated in suspected neurological manifestations associated with HEV infection. In this case always provide a paired sample (plasma or serum) to evaluate intrathecal synthesis of antibodies.
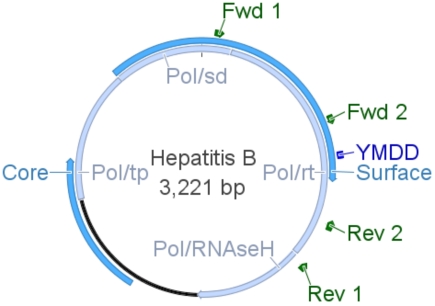
The genetic organization of HBV is extremely compact with part of the core and all of the surface genes (blue) overlapping the polymerase gene (light blue). Consequently, mutations affecting one gene product may affect the other. The polymerase is subdivided into functional domains: terminal protein (tp), spacer (sd), reverse transcriptase (rt) and RNAseH. The active site of rt (YMDD, dark blue) and adjacent sequences are the target of mutations conferring resistance to antiviral drugs. The core protein constitutes the viral capsid, while the surface antigen constitutes the envelope of the virus upon anchoring in a lipid membrane of cellular origin. Other important features are not represented here for sake of simplicity: the pregenomic RNA used as template for viral DNA replication, the X open reading frame whose function is unknown as well as regulatory elements necessary for viral gene expression and viral DNA replication. Binding sites for primers used in diagnostics are shown in green.
MOLECULAR TOOLS TO IDENTIFY MUTATIONS
The identification of mutations can only be done after polymerase chain reaction (PCR) amplification of rt with DNA extracted from the plasma of infected patients. Primers used in our laboratory are depicted in green on the HBV genome displayed above.
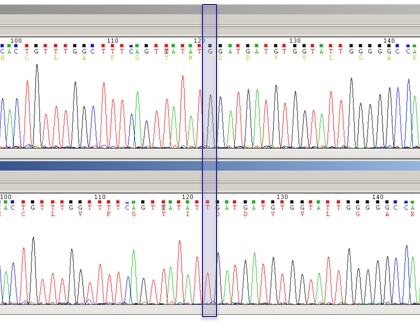
Precise identification of mutations can be done by reverse blotting hybridization against a panel of probes (for known mutations, high sensitivity of detection), or by DNA sequencing (for known as well as for unknown mutations) provided they represent at least 20% of the whole population. This level of detection is sufficient for routine diagnostics. Once the DNA sequence is determined, it is translated into its corresponding amino acid stretch. Mutations can then be identified after alignment of the amino acid sequence against reference HBV protein databases. This method, used in our laboratory, is illustrated below by sequencing chromatograms of two samples from the same patient, either before (upper strand) or after failure treatment with telbivudine (lower strand). The G/T transversion indicated by the blue shade leads to the M204I mutation known to confer resistance to this drug.
HDV RNA testing
The genetic information of HDV is carried by a circular RNA molecule with extensive base pairing. This structure poses a challenge for PCR techniques aimed at the quantitative assessment of HDV viremia due to intramolecular base pairing competing with primer binding. To overcome this difficulty, we use a two-step reverse transcription real-time PCR and a rapid PCR cycling profile. This assay has been validated against known samples, and is a very good performer in a recently established European Quality Control Scheme.
HEV RNA testing
We have adpated a broad range real-time PCR assay published by Jothikumar et. al. (2006, J. Virol. Methods 131(1): 65-71.) to overcome the high genetic variability of HEV. The primers/probe are regularly monitored for matching most strains deposited in GenBank, covering the 4 known genotypes. It is best to send samples as early as possible after symptom manifestation for utmost clinical sensitivity.