Hepatitis B Resistance Testing
THE HEPATITIS B VIRUS
The hepatitis B virus (HBV) is a member of the Hepadnaviridae, a family of DNA viruses which is unique in that viral DNA replication requires an RNA intermediate; hence the viral polymerase possesses reverse transcriptase activity like that found in retroviruses. This feature is associated with the genetic variability of HBV and persistent infection. There are presently 8 HBV genotypes (A-H) based on sequence divergence of their 3.2 kb circular genome.
THE HBV LIFE CYCLE: BASIS FOR PERSISTENT INFECTION
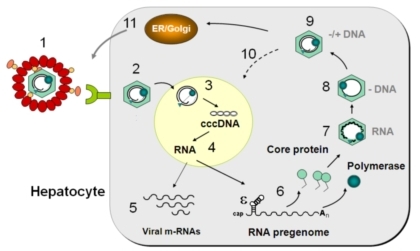
The enveloped virus particle attaches (1) to the hepatocyte through recognition of a membrane-anchored cellular receptor. After its translocation to the cytoplasm (2), the nucleocapsid releases the viral genome to the nucleus (3) where it is converted into a covalently-closed circular DNA (cccDNA) to serve as template for viral RNA transcription (4). The viral mRNAs (5) are translated into viral proteins in the cytoplasm, and the RNA pregenome (6) is encapsidated together with the viral polymerase into a capsid made of core protein (7). Upon maturation of the virus particle (8-9), the viral DNA is synthesized by the viral polymerase using the encapsidated pregenome as template. The newly synthesized nucleocapsid can either release the viral DNA in the nucleus initiating a new round of cccDNA synthesis, maintaining for a long time the viral genome in the infected cell and increasing the amount of viral RNAs (10), or acquire the viral envelope in the endoplasmic reticulum (ER) and Golgi apparatus with release of mature virions from infected cell by exocytosis (11). This mode of virus production does not result in cell death, thus contributing to chronic infection of the liver.
EMERGENCE OF ANTIVIRAL RESISTANCE
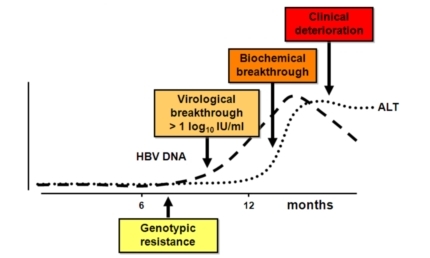
Like with retroviruses, the reverse transcriptase activity of the viral polymerase incorporates wrong nucleotides in the nascent DNA chain at a much higher frequency compared to cellular DNA synthesis. This results in a population of viral genomes with a high proportion of variants forming a quasispecies in the infected patient, and from which resistant viruses can arise. Genotypic resistance is defined by the emergence of HBV variants with mutations in rt that render the polymerase deficient at incorporating the nucleotide analogs used to treat the patient. Replication of the viral DNA in mutant infected cells is therefore much less sensitive to drugs, and virus production increases despite treatment. Virological breakthrough is reached when viral DNA load is superior to one log the baseline level. This level of virus replication precedes the alteration of liver functions, as measured by increasing alanine transferase (ALT) concentrations (biochemical breakthrough), and clinical deterioration. Thanks to the identification of viral mutations characteristic of genotypic resistance, one can adapt treatment of patients early enough to prevent biochemical breakthrough and overt disease.
HBV MUTATIONS FREQUENTLY INVOLVED IN RESISTANCE TO ANTIVIRALS
Well known mutations associated with drug resistance are reported above. HBV resistance to antivirals is due to mutations concentrated in the rt domain of the polymerase, near the YMDD peptide motif found at the active site of rt. Mutations are numbered according to the amino acid position within the rt domain. The codon for Methione (M) at position 204 is a key target for mutations changing M to Isoleucine (I) or to Valine (V). These mutations are reported as M204I or M204V respectively. While M204I can be found isolated in patients infected with LAM resistant viruses, M204V is always accompanied by another mutation like L180M. Such a mutation is necessary to compensate the replication defect induced by M204V, thus improving the fitness of the variant virus in the presence of the antiviral drug.
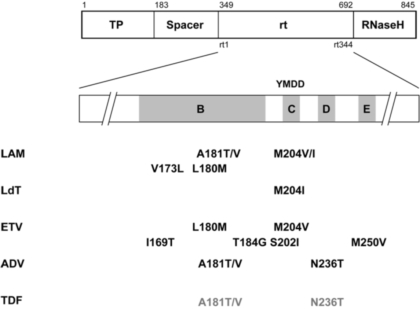
LAM: lamivudine; ADV: adefovir; ETV: entecavir; LdT: telbivudine; TDF: tenofovir. Note that the mutations indicated in grey reduce the susceptibility of HBV to TDF in vitro but that virological breakthrough has not been documented yet in patients.



