Dominique Sanglard Lab
Research Topics
Fungal infections in human are still causing high mortality worldwide, even if of antifungal agents are used to fight these diseases. Among existing fungal pathogen, Candida species are the most frequent and in particular C. albicans and C. glabrata. These two fungal species possess specific virulence traits and also develop resistance to currently available antifungal agents.
Our research group investigates both fungal species in order to better understand the features that contribute to their virulence. For this purpose, advanced genetic and molecular tools are used as well as several animal models of infection. Our studies also have a focus on the molecular understanding of antifungal resistance mechanisms from fungal samples in clinical cases.
In addition to these two major activities, our research group is trying to identify novel antifungal agents from different sources and attempt to establish their activities directly in vivo using animal models of infection.

Impact of antifungal resistance in in vivo fitness of fungal pathogens
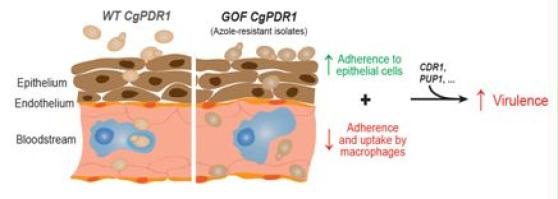
Resistance mechanisms to azoles, which are agents widely used in therapy, may have negative effects on the fitness of fungal pathogen in their hosts. We are investigating these effects with animal models using Candida albicans and Candida glabrata that are two important fungal pathogens and into which azoles resistance mechanisms are well understood. Our results showed that, while azole resistance has either neutral or negative effects on fitness in C. albicans, azole resistance in C. glabrata generally provides beneficial virulence traits. Our recent results point towards enhanced expression of adhesins that is coupled with azole resistance as a principal cause of the gain of fitness in infection models.
Funding : FNRS 31003A_146936 (2013-2016)
Molecular basis of antifungal drug tolerance in Candida albicans.
Antifungal resistance mechanisms have been extensively described in C. albicans for available antifungal agents. Antifungal tolerance operates at antifungal concentrations above individual intrinsic inhibitory values. Tolerance to antifungal agents favours the emergence of persister cells, which are able to survive antifungal therapy and can cause relapses. We are currently evaluating the effect of antifungal tolerance in the efficacy of drug treatments in animal models and also investigating the molecular basis of antifungal tolerance.
Funding: FNRS 31003A_146936 (2013-2016)
Novel genome-wide transcriptomic approaches to challenge Candida albicans-hosts interactions (Sinergia)
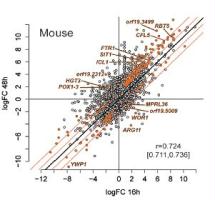
This project aims to use several genomic approaches in order to discover novel cellular factors involved in the interaction of the human pathogen Candida albicans with cells of the host. The project is composed of three different complementary research partners, each of which will contribute to different aspects of the project. The first two partners (D. Sanglard and S. Leibundgut, ETHZ) will contribute to the understanding of fungal and host factors in the fungal-host interactions, respectively. The third partner (M. Pagni, University of Lausanne) will be associated with the genomic data analysis and development of bioinformatic tools.
Funding: FNRS CRSII3_141848 (2013-2017)
Identification of novel modes of action of plant natural products targeting fungal human and phytopathogens
Fungi contribute to beneficial associations for human and in the environment, however they are also the cause of a wide spectrum of diseases. Due to the few antifungal availlable and occurrence of antifungal resistance , it is necessary to identify novel sources of effective agents. We propose to address the identification of antifungal agents from natural sources originating from plants. Natutral products are isolated by the laboratory of J.-L. Wolfender (University of Geneva) and screened for antifungal activity. Interesting and novel compounds are interrogated for their spectrum of activity, mode of action and efficacy in animal models.
Funding: FNRS CR23I3_143733 (2013-2016)

Host-fungus cross-talk in the emergence of resistance and in the success of infection
Candida albicans is an opportunistic pathogen causing oral, vaginal or systemic infections in immuno-compromized patients. In up to 30% of the patients with C. albicans systemic infections, the issue is fatal despite existing treatments. C. albicans exposed for a long time to antifungals adapts to this stress, and develops resistance representing one of the causes of treatment failure. Therefore, alternative treatment strategies are needed.
- Study of host-pathogen interaction, a key component of any infectious process. Treatment modifying this relationship can be designed. Transcription factors (TF) are potentially important for C. albicans virulence since they integrate several signals from host environment and participate in an adapted microbial response. In this project, we developed an original approach to analyze a collection of C. albicans transcription factor (TF) mutant strains in vivo in order to identified TF crucial for virulence. Interesting candidates are now analysed.
- Deciphering factors influencing antifungal resistance emergence in vivo in multi-species population infection. We are thus developing a system of in vivo experimental development of resistance. Factors such as animal environment (mice or Galleria mellonella versus in vitro conditions), or clonal interference due to co-infecting C. albicans strains on a given studied strain, are analysed.
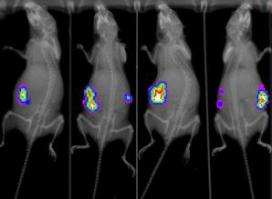
- Development of bioluminescence tools to follow infection in vivo, in mice or Galleria mellonella over time. To be able to follow infection and eventually emergence of resistance, we first developed a bioluminescent detection of C. albicans in G. mellonella living larvae using a common CCD camera. Then we are also optimizing bioluminescence reporter tools to follow infection in mice using a recently acquired optical iIn-vivo imaging system X-treme II (Bruker). This will allow a better characterization of the infection of two different hosts the mouse and the wax moth G. mellonella. In mice, we will monitor emergence of resiatnce using a bioluminescence reporter switched-on thanks to acquisition of gain-of-function mutations in antifungal resistance genes.
Funding: Novartis Foundation for biological and medical research.